April 17, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Extrachromosomal DNA detection in Barrett's esophagus linked to cancer development
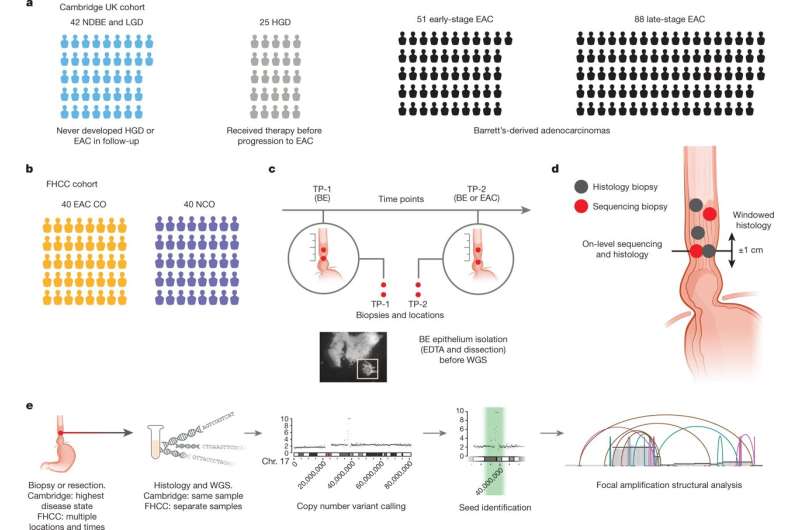
A multi-institution study that included researchers from Stanford University, the University of California at San Diego, the University of Cambridge, the Fred Hutchinson Cancer Center and others has looked into the development of extrachromosomal DNA in patients with esophageal adenocarcinoma or Barrett's esophagus.
In the paper, "Extrachromosomal DNA in the cancerous transformation of Barrett's oesophagus," published in Nature, researchers identify previously unknown aspects of extrachromosomal DNA presence and their potential role in Barrett's esophagus. David H. Wang has published a News & Views piece in the same journal discussing the study.
Barrett's esophagus is a pre-cancerous tissue abnormality that affects about 1.6% of the U.S. population. The condition is mostly harmless, defined by cells in the esophagus lining that become more intestinal-like and is frequently associated with heartburn and acid regurgitation.
While potentially cancerous, identification of the disease is followed up by frequent monitoring and only treated if there are substantial progressive changes to the esophagus lining. The good news is that most people diagnosed with Barrett's esophagus (around 92%) do not develop esophageal adenocarcinoma (EAC). This, unfortunately, also means that around 8% of patients with Barrett's esophagus will develop EAC.
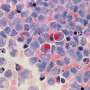
Mutated gene amplification on extrachromosomal DNA (ecDNA) has been previously understood to drive the evolution of tumors and treatment resistance and is associated with poor outcomes. It has been unclear whether ecDNA is a later manifestation of genomic instability of tumors, or an early occurrence in the transition from abnormal tissue to cancer, though it has been expected to be a later based on previous observational studies.
To investigate, researchers analyzed whole-genome sequencing data from patients with esophageal adenocarcinoma or Barrett's esophagus. These data included 206 biopsies in Barrett's esophagus surveillance and EAC cohorts from Cambridge University.
Researchers also analyzed whole-genome sequencing and histology data from biopsies collected across multiple regions at two time points from 80 patients in a case-control study at the Fred Hutchinson Cancer Center.
In the Cambridge cohorts, the frequency of ecDNA increased between Barrett's esophagus early-stage (24%) and late-stage (43%) EAC, suggesting that ecDNA is formed during cancer progression.
In the Fred Hutchinson Cancer Center cohort, 33% of patients who developed EAC had at least one esophageal biopsy with ecDNA before or at the diagnosis of EAC.
In biopsies collected before cancer diagnosis, higher levels of ecDNA were present in samples from patients who later developed EAC than in samples from those who did not develop cancer. The early presence of ecDNA ruled out the scenario where ecDNA comes about as a later tumor-related manifestation of genome instability.
The findings are hugely significant in that they reveal that ecDNA can arise before the development of diagnosed cancer and that higher levels are associated with progression. This suggests the possibility of a pre-cancerous disease marker, allowing for clinically informed earlier interventions for Barrett's esophagus patients by conducting ecDNA testing at early disease presentation. More research is needed to develop clinical detection and treatment methods.
More information: Jens Luebeck et al, Extrachromosomal DNA in the cancerous transformation of Barrett's oesophagus, Nature (2023). DOI: 10.1038/s41586-023-05937-5
David H. Wang, Extrachromosomal DNA appears before cancer forms, Nature (2023). DOI: 10.1038/d41586-023-00982-6
© 2023 Science X Network