This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Enhanced recombination among omicron variants shown contributes to viral immune escape, finds study
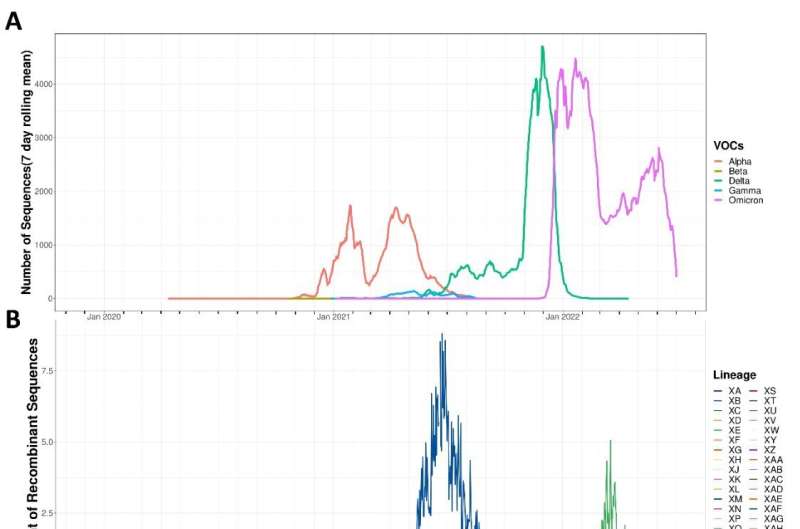
In January 2022, around the time that the omicron variant of SARS-CoV-2 started spreading rapidly, a team of researchers at the Indian Institute of Science (IISc) led by Shashank Tripathi, Assistant Professor at the Department of Microbiology & Cell Biology, and Centre for Infectious Diseases Research, noticed that there was an unusually high increase in the number of recombinant strains of the omicron variant.
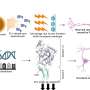
The team analyzed genomic sequences of all the viral strains that appeared between November 2019 and July 2022 in various databases worldwide. In a study published in the Journal of Medical Virology, they have identified several new mutations that accumulated through recombination at a high rate and affected viral proteins, especially different parts (domains) of the viral spike protein.
These domains—such as the Receptor Binding Domain (RBD) and N Terminal Domain (NTD)—are known to be involved in the virus-host binding and have also been reported as sites of attack by the host's immune system. The team showed that, with the aid of these mutations, several such omicron recombinant and mutant strains were able to escape from the host's defenses and bind more tightly to the host cell.
Their observations add to growing evidence about how efficient new strains of the virus are at escaping immune attack and at causing infections.
Viruses like SARS-CoV-2 are known to change constantly, making it hard for our immune system to identify and destroy them. This is a major concern when generating vaccines. The genetic material in SARS-CoV-2 is a long single-stranded RNA. In addition, the protein that is required to make copies of this RNA—RNA polymerase—is known to be error-prone in this virus.
Viruses can evolve via one of two mechanisms: mutation or recombination, explains Tripathi. "This is a strategy to increase its genetic diversity." The polymerase doesn't just allow mutations to accumulate, it often also causes recombination to happen between different strains of the virus. This is possible when there is co-infection—when a host cell is infected by more than one strain of the virus.
"When copying the viral RNA, the polymerase can jump from one RNA template to another that is nearby," says Tripathi. If the nearby sequence is that of another strain, then the new copy will be a recombinant or a hybrid of the two parental strains. Tripathi says that there are currently more than 35 recombinants of SARS-CoV-2. For example, one of the more efficient variants, XBB, which emerged in 2022, was born from recombination between two other versions of omicron, he explains.
There are two possible reasons for the increase in these recombination events, according to the study. First, the number of infections and co-infections were high during the 2022 omicron wave. Second, the team noticed that a specific mutation has appeared in a viral gene for an exonuclease, a protein that can cleave RNA and is believed to be involved in recombination. Tripathi explains, "Our findings show that the virus is not cooling down but is actually warming up as far as mutations go."
Because enhanced recombination can increase the chances of new strains emerging, tracking such recombinations through regular sequencing of the virus is crucial, the researchers say.
More information: Rishad Shiraz et al, Enhanced recombination among Omicron subvariants of SARS‐CoV‐2 contributes to viral immune escape, Journal of Medical Virology (2023). DOI: 10.1002/jmv.28519