This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study shows new route for dangerous coronavirus strain emergence
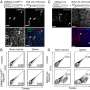
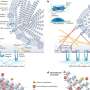
![Mutational escape from cytotoxic T cells. a Calculation of site presentation scores (adapted from Marty et al). b, c Change of PHBR scores caused by mutations for HLA I b and HLA II c, respectively. Dot color corresponds to PHBR fold change; the mutations that substantially (>3-fold) increase PHBR are signed. Sites that did not bind any of the patient’s HLA alleles both in ancestral and derived states are not shown. d Comparison of imBR scores for the mutated sites in their ancestral and derived states (n = 11). The level of significance is calculated by the two-sided paired Wilcoxon exact sign-rank test: Test statistic V = 0, p-value = 0.0009766, 95% confidence interval: [−11.7835; −0.3065], median estimates −4.647. Horizontal lines in a boxplot represent minima, first quartile, median, third quartile, and maxima. Credit: Nature Communications (2023). DOI: 10.1038/s41467-022-34033-x Study shows new route for dangerous coronavirus strain emergence](https://scx1.b-cdn.net/csz/news/800a/2023/study-shows-new-route.jpg)
The immune system fights the coronavirus with antibodies and T cells. Scientists have a fairly good idea of how the virus avoids antibodies, including those promoted by vaccination, which is how variants of concern such as the omicron are known to appear. T cell evasion, on the other hand, has remained poorly understood.
Now, Skoltech researchers and their colleagues report mutations enabling T cell escape, which emerged in a patient with compromised immunity. According to their paper in Nature Communications, this is a new source of dangerous mutant strains.
For antibodies and T cells to work, they need to be able to hold on to the viral particle using what's known as an antigen. This refers to some constituent molecule, such as a protein or peptide, that characterizes that pathogen. Antigens are displayed (made accessible) to the immune system by specialized antigen-presenting molecules. Now, the pesky virus itself is constantly evolving, leading to shape-shifts that may alter the region containing the antigen, making it hard or impossible to grab.
Keeping track of mutations that allow SARS-CoV-2—the virus causing COVID-19—to avoid the immune response is crucial for understanding, monitoring, and controlling the pandemic. But while the mutants capable of evading antibodies are largely on the scientists' radar and in the news, we have remained in the dark about viral adaptations to T cell immunity.
"To see if the virus evolves T cell escape, we analyzed the case of a patient who had COVID for 318 days. She was simultaneously receiving cancer therapy that suppresses antibody production as a side effect. This allowed us to observe T cell response and viral adaptation to it in isolation," Professor Georgii Bazykin of Skoltech Bio, the principal investigator of the study, commented. "And indeed, bioinformatic analysis revealed that over the course of this lengthy disease, the virus accumulated precisely those mutations that prevented the patient's T cells from recognizing it."
He explained that the researchers know this because every person's T cell immunity is different. The way T cells work is they go over the entire set of "keys" available to them until they can sink one of them into the "lock"—that is, the surface of the antigen displayed by antigen-presenting molecules. What happens is the lock keeps shape-shifting, and at some point no key fits and the mutation that brought this about carries on.
Moreover, viral mutations can complicate finding the right key or allow the antigen to avoid being paraded by the presenting molecules. Such mutations are advantageous to the virus and are therefore perpetuated by natural selection. Since both the set of keys (T cell receptors) and the presenting molecules vary from person to person, researchers could demonstrate that the virus in the patient actually evolved adaptations to her immunity as opposed to acquiring mutations at random.
Now, are such adaptations a concern for individual long COVID patients with compromised immunity, or are they a potential global threat? To settle this, the researchers analyzed to what extent resistance to T cells with the particular set of presenting molecules characteristic for the studied patient also protected the virus from the entire diversity of T cell response represented by the global population. It turned out that the T cell escape mutations discovered were indeed of a kind that would help the virus invalidate many of humanity's most widespread antigen-presenting molecules.
The findings make all the more relevant the concern that when the virus is left to its own devices for a long time in a patient, it is likely to evolve uncanny mutations. If the new dangerous strain then escapes the patient by infecting other individuals, this could trigger the rise of a new variant of concern. And we now know strains resistant to T cell-based immunity could emerge that way, too, in addition to those evading antibodies produced in response to vaccination or natural infection.
"So far it is hard to judge to what extent such potential mutant strains are dangerous, but we will keep investigating the matter to find out," Bazykin said.
More information: Oksana V. Stanevich et al, SARS-CoV-2 escape from cytotoxic T cells during long-term COVID-19, Nature Communications (2023). DOI: 10.1038/s41467-022-34033-x