This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Machine learning model finds genetic factors for heart disease
To get an inside look at the heart, cardiologists often use electrocardiograms (ECGs) to trace its electrical activity and magnetic resonance images (MRIs) to map its structure. Because the two types of data reveal different details about the heart, physicians typically study them separately to diagnose heart conditions.
Now, in a paper published in Nature Communications, scientists in the Eric and Wendy Schmidt Center at the Broad Institute of MIT and Harvard have developed a machine learning approach that can learn patterns from ECGs and MRIs simultaneously, and based on those patterns, predict characteristics of a patient's heart. Such a tool, with further development, could one day help doctors better detect and diagnose heart conditions from routine tests such as ECGs.
The researchers also showed that they could analyze ECG recordings, which are easy and cheap to acquire, and generate MRI movies of the same heart, which are much more expensive to capture. And their method could even be used to find new genetic markers of heart disease that existing approaches that look at individual data modalities might miss.
Overall, the team said their technology is a more holistic way to study the heart and its ailments. "It is clear that these two views, ECGs and MRIs, should be integrated because they provide different perspectives on the state of the heart," said Caroline Uhler, a co-senior author on the study, a Broad core institute member, co-director of the Schmidt Center at Broad, and a professor in the Department of Electrical Engineering and Computer Science as well as the Institute for Data, Systems, and Society at MIT.
"As a field, cardiology is fortunate to have many diagnostic modalities, each providing a different view into cardiac physiology in health and diseases. A challenge we face is that we lack systematic tools for integrating these modalities into a single, coherent picture," said Anthony Philippakis, a senior co-author on the study and chief data officer at Broad and co-director of the Schmidt Center. "This study represents a first step towards building such a multi-modal characterization."
Model making
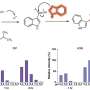
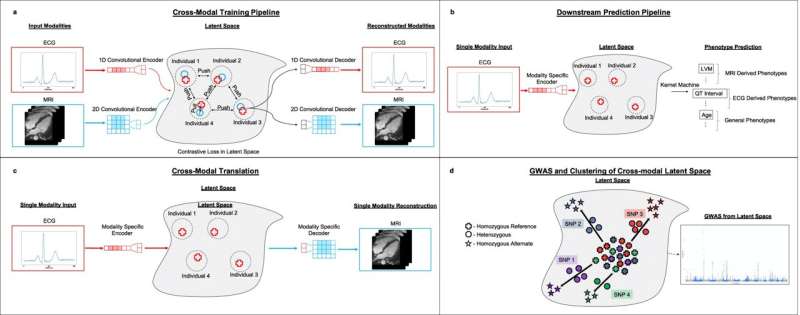
To develop their model, the researchers used a machine learning algorithm called an autoencoder, which automatically integrates gigantic swaths of data into a concise representation—a simpler form of the data. The team then used this representation as input for other machine learning models that make specific predictions.
In their study, the team first trained their autoencoder using ECGs and heart MRIs from participants in the UK Biobank. They fed in tens of thousands of ECGs, each paired with MRI images from the same person. The algorithm then created shared representations that captured crucial details from both types of data.
"Once you have these representations, you can use them for many different applications," said Adityanarayanan Radhakrishnan, a co-first author on the study, an Eric and Wendy Schmidt Center Fellow at the Broad, and a graduate student at MIT in Uhler's lab. Sam Friedman, a senior machine learning scientist in the Data Sciences Platform at the Broad, is the other co-first author.
One of those applications is predicting heart-related traits. The researchers used the representations created by their autoencoders to build a model that could predict a range of traits, including features of the heart like the weight of the left ventricle, other patient characteristics related to heart function like age, and even heart disorders. Moreover, their model outperformed more standard machine learning approaches, as well as autoencoder algorithms that were trained on just one of the imaging modalities.
"What we showed here is that you get better prediction accuracy if you incorporate multiple types of data," Uhler said.
Radhakrishnan explained that their model made more accurate predictions because it used representations that had been trained on a much larger dataset. Autoencoders don't require data that have been labeled by humans, so the team could feed their autoencoder with around 39,000 unlabeled pairs of ECGs and MRI images, rather than just around 5,000 labeled pairs.
The researchers demonstrated another application of their autoencoder: generating new MRI movies. By inputting an individual's ECG recording into the model—without a paired MRI recording—the model produced the predicted MRI movie for the same person.
With more work, the scientists envision that such technology could potentially allow physicians to learn more about a patient's heart health from just ECG recordings, which are routinely collected at doctors' offices.
Broader gene search
With their autoencoder representations, the team realized they could also use them to look for genetic variants associated with heart disease. The traditional method of finding genetic variants for a disease, called a genome-wide association study (GWAS), requires genetic data from individuals that have been labeled with the disease of interest.
But because the team's autoencoder framework doesn't require labeled data, they were able to generate representations that reflected the overall state of a patient's heart. Using these representations and genetic data on the same patients from the UK Biobank, the researchers created a model that looked for genetic variants that impact the state of the heart in more general ways. The model produced a list of variants including many of the known variants related to heart disease and some new ones that can now be investigated further.
Radhakrishnan said that genetic discovery could be the area in which the autoencoder framework, with more data and development, could have the most impact—not just for heart disease, but for any disease. The research team is already working on applying their autoencoder framework to study neurological diseases.
Uhler said this project is a good example of how innovations in biomedical data analysis emerge when machine learning researchers collaborate with biologists and physicians. "An exciting aspect about getting machine learning researchers interested in biomedical questions is that they might come up with a completely new way of looking at a problem."
More information: Adityanarayanan Radhakrishnan et al, Cross-modal autoencoder framework learns holistic representations of cardiovascular state, Nature Communications (2023). DOI: 10.1038/s41467-023-38125-0