This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Genome-wide CRISPR screens identify PARP inhibitor sensitivity and resistance in prostate cancer
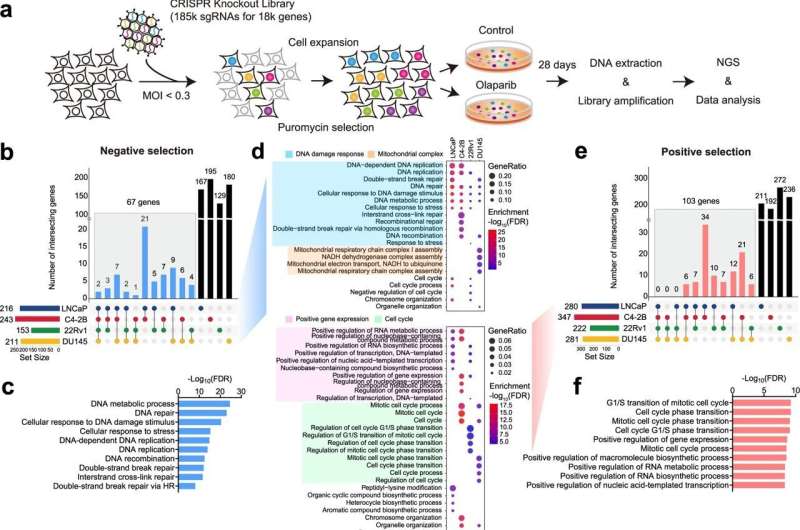
Prostate cancer tumors harboring BRCA1/2 mutations are exceptionally sensitive to PARP inhibitors, while genomic alterations in other DNA damage response (DDR) genes are less responsive. To identify previously unknown genes whose loss has a profound impact on PARP inhibitor response, researchers from Dana-Farber Brigham Cancer Center led a multinational effort to perform genome-wide CRISPR-Cas9 knockout screens. The study goal was to inform the use of PARP inhibitors beyond BRCA1/2-deficient tumors and support reevaluation of current biomarkers for PARP inhibition in prostate cancer.
The findings are published in the journal Nature Communications.
The study identified multiple novel genes (e.g., MMS22L and RNASEH2B) that are frequently deleted in prostate cancer. These genes could serve as predictive biomarkers for PARP inhibitor response in prostate cancer, according to the study. The research team also found that loss of CHEK2 (the FDA-approved biomarker for therapeutic response to olaparib) confers resistance, rather than sensitivity, to PARP inhibition.
"Current targeted cancer therapies, including PARP inhibitors, are largely guided by mutations of a single gene and overlook concurrent genomic alterations," said Li Jia, Ph.D., director of Urology Research, Brigham and Women's Hospital. "We found that PARP inhibitor sensitivity instead may depend on interaction between multiple genomic alterations. Therefore, comprehensive genomic analysis may help improve clinical decision making."
More information: Takuya Tsujino et al, CRISPR screens reveal genetic determinants of PARP inhibitor sensitivity and resistance in prostate cancer, Nature Communications (2023). DOI: 10.1038/s41467-023-35880-y